Portfolio
Below are a few examples of visuals from various analyses. These works are not exhaustive and represent only a small sample of completed projects. The graphs presented here are intentionally taken out of context and without labels, for confidentiality reasons (except for the work on BRCA2)
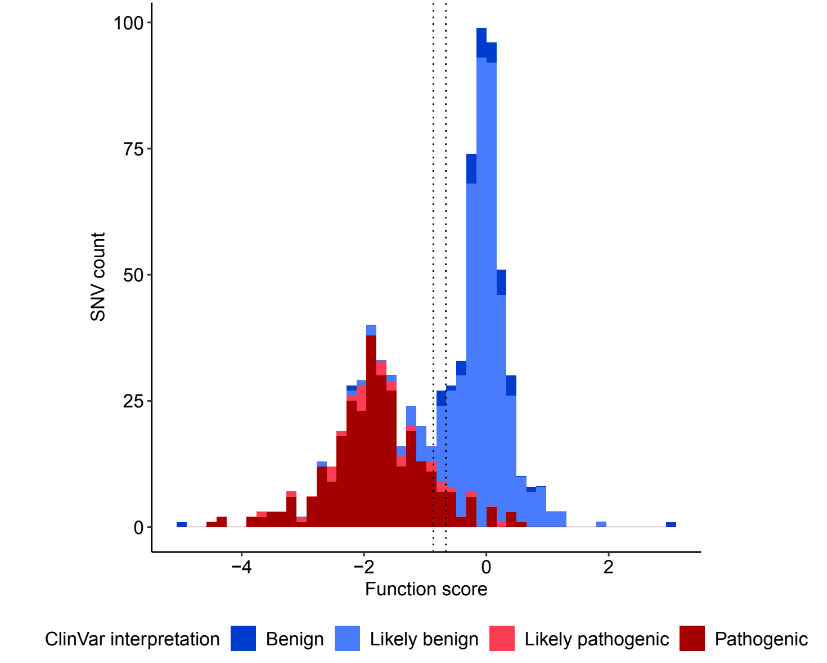
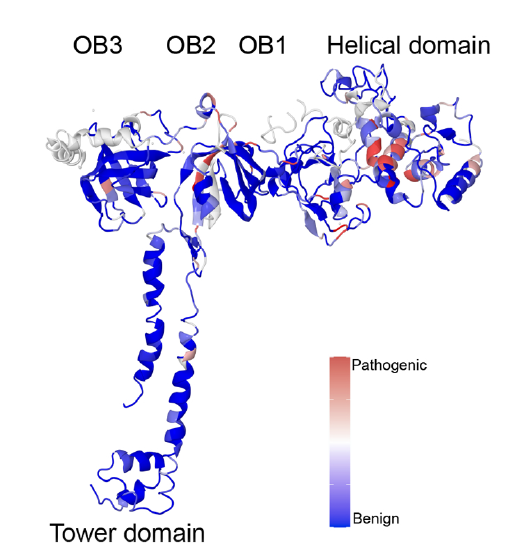
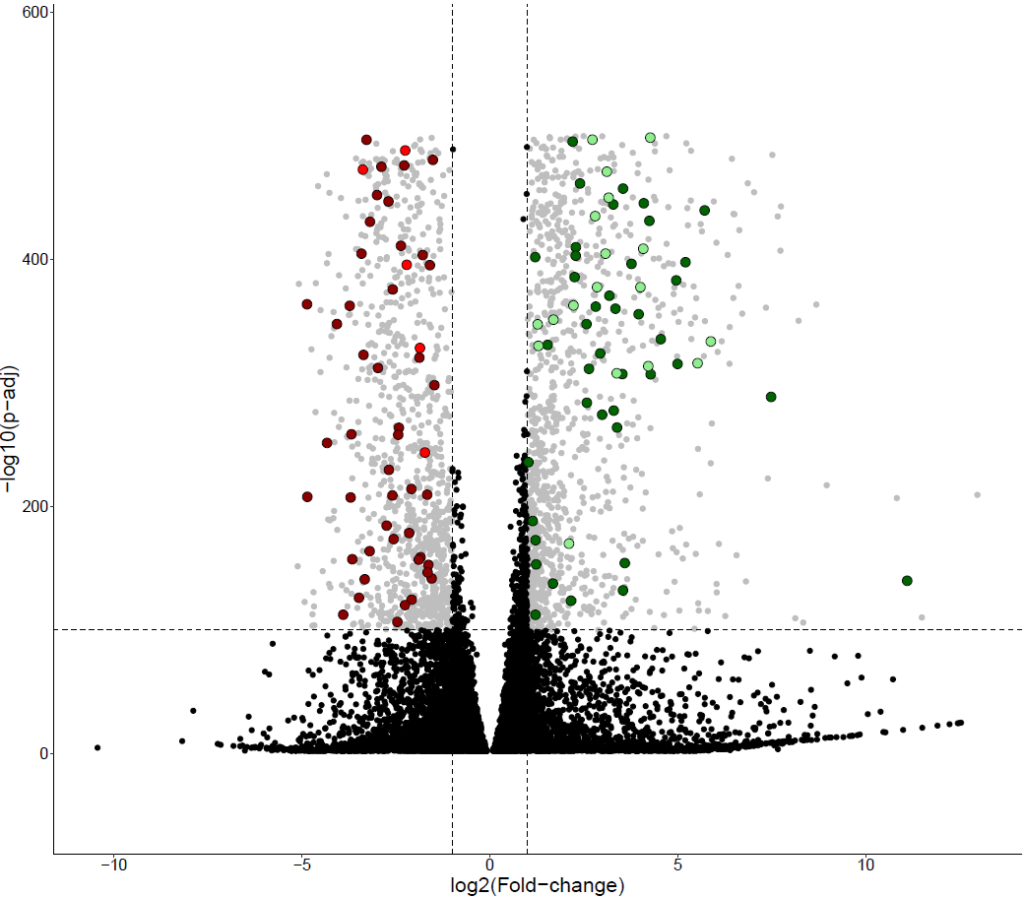
Clinical classification of BRCA2 variants through genome-wide saturation
The objective of this project is to perform a functional test of the BRCA2 gene based on CRISPR-Cas9. All possible SNVs have been generated across several exons of the gene. By using cell survival data, the goal is to predict the pathogenicity of these variants, most of which have never been studied before.
To achieve this, we developed an automated pipeline for processing the (numerous) data, created and refined the prediction model. A R Shiny application was also developed
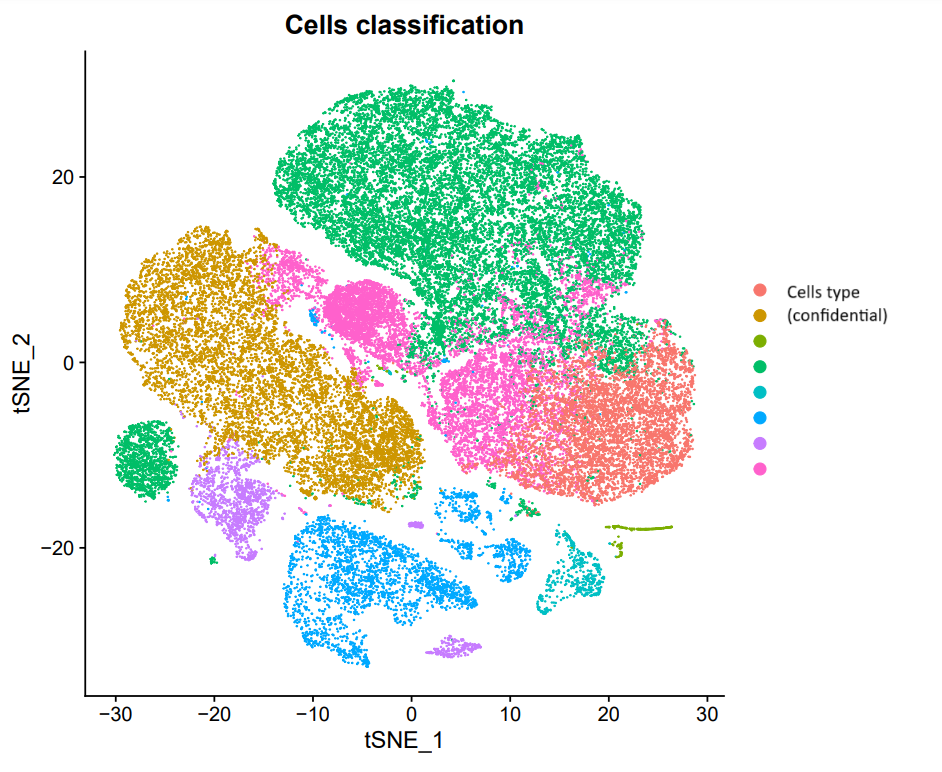
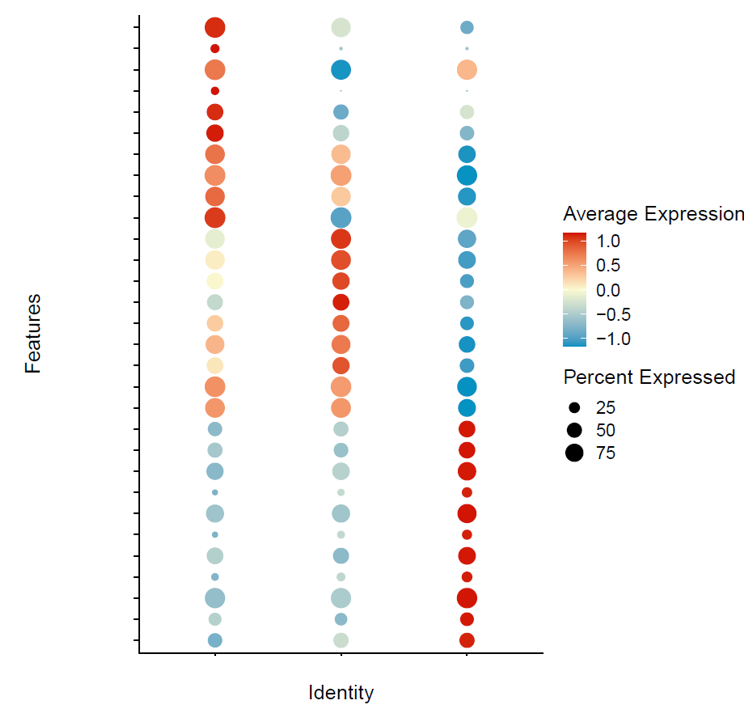
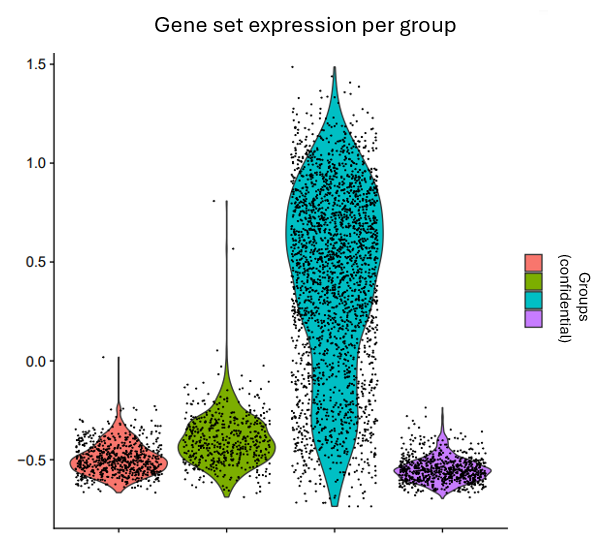
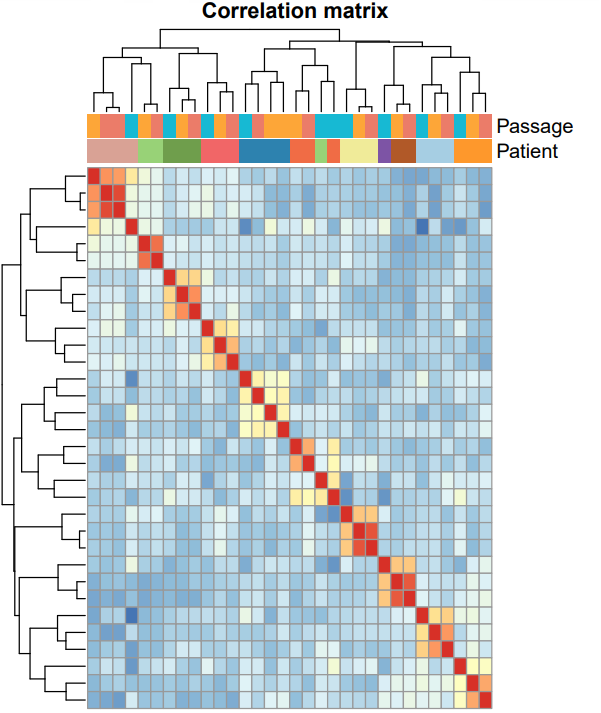
scRNA-seq analyses
scRNA-seq analyses including data preparation, cell annotation, differential expression analysis, marker studies, pseudotime analysis, pathway analysis, etc. Additionally, stRNA-seq (spatial transcriptomics) analysis with annotation of different cell groups in correlation with histology.
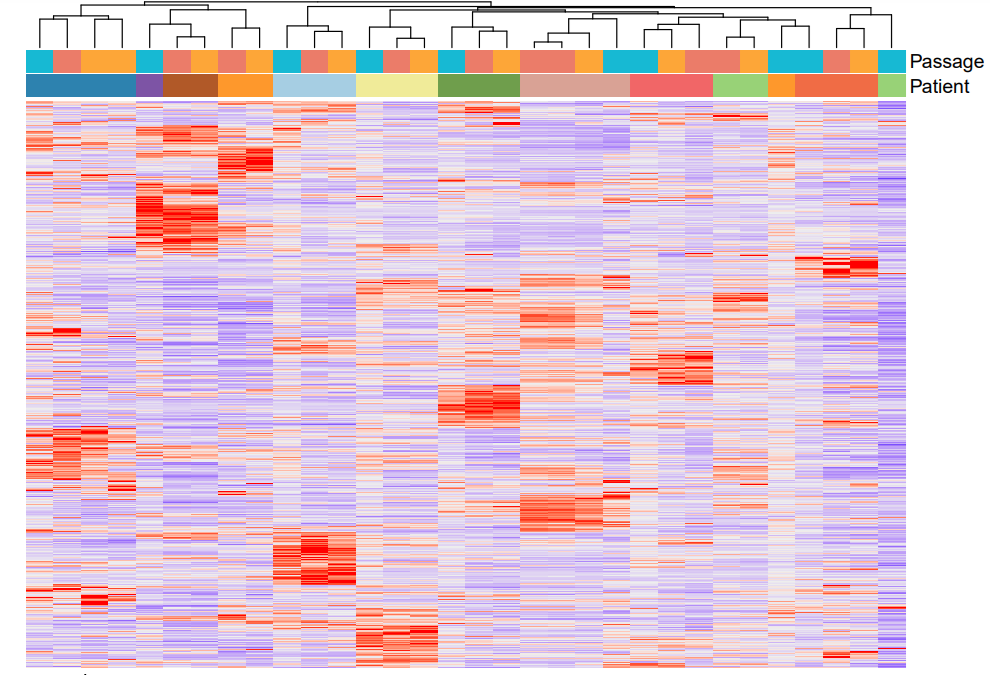
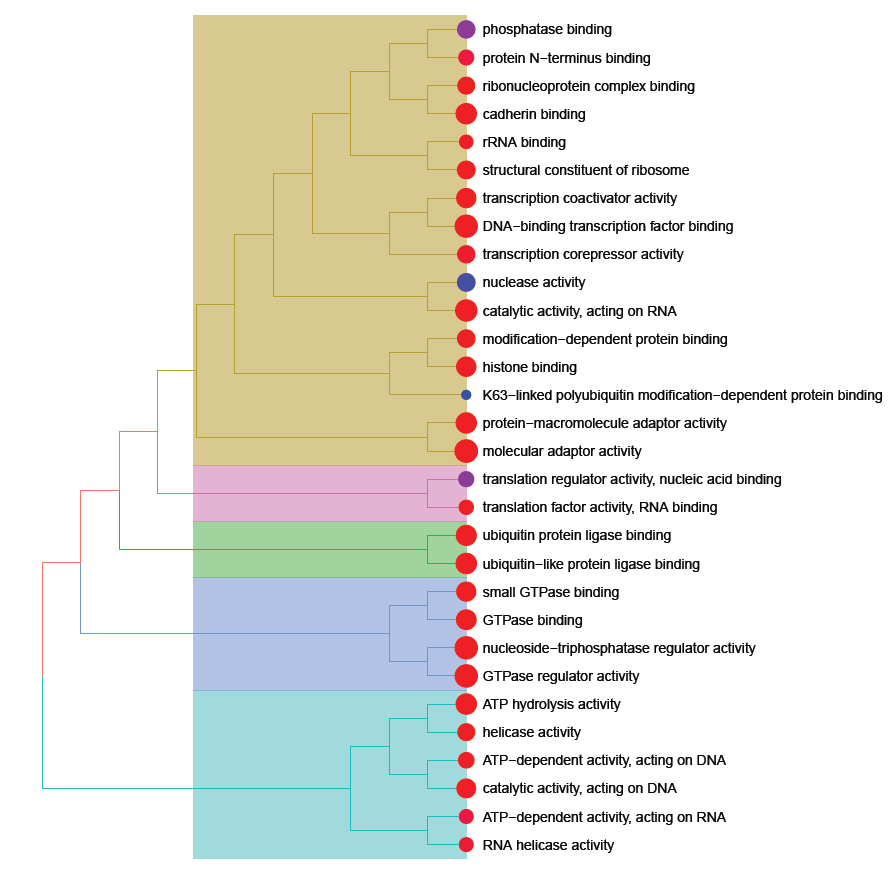
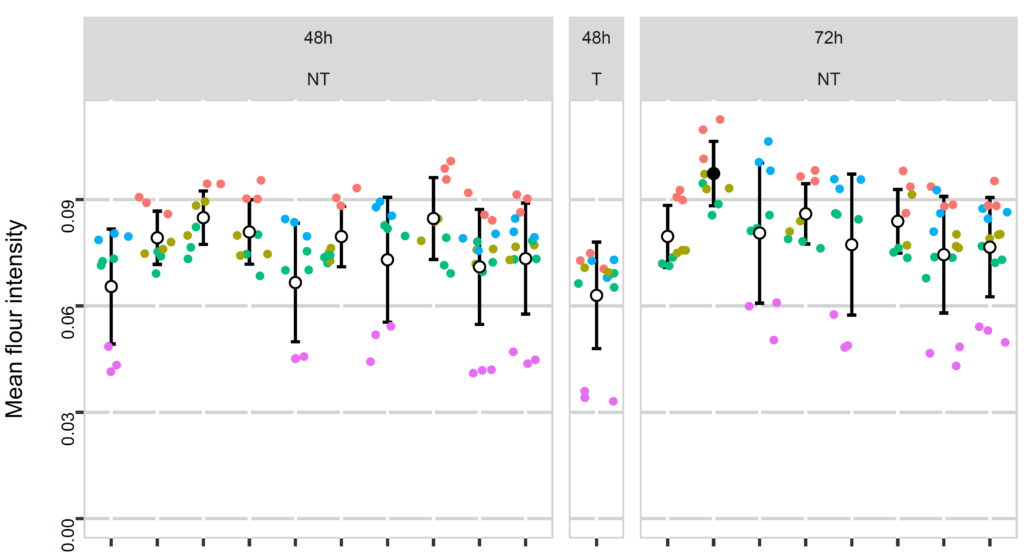
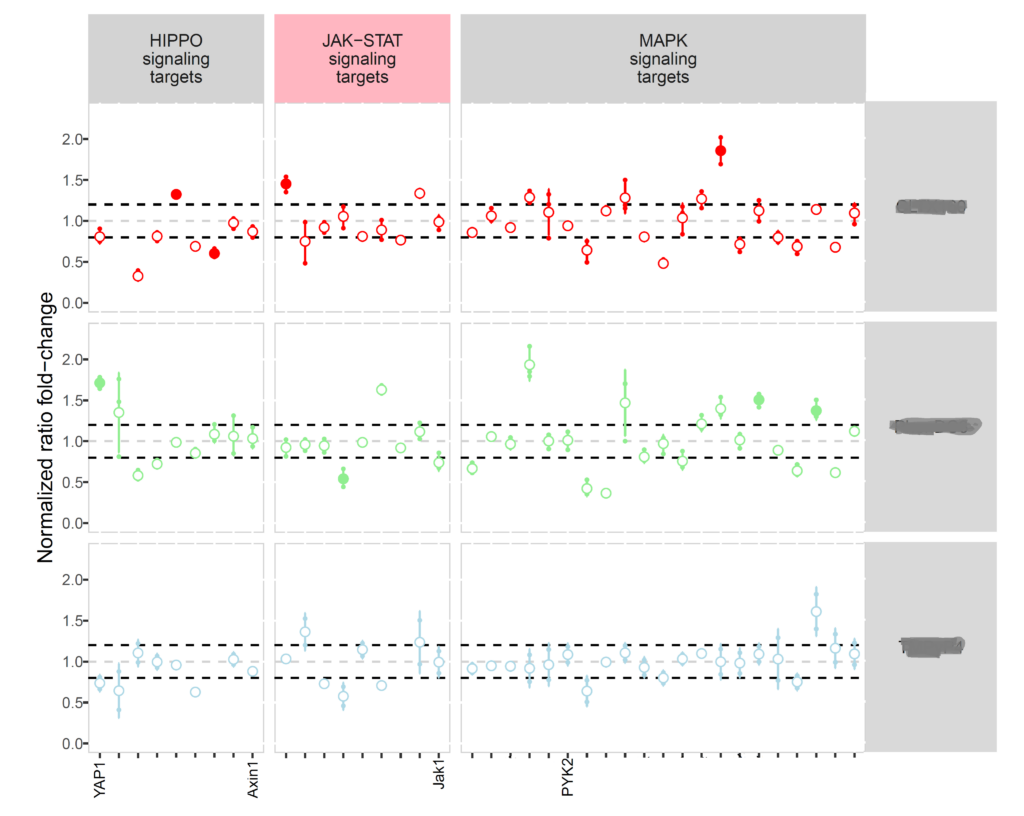
RNA-seq analyses
Various RNA-seq analyses: differential expression analysis, isoform study, correlation between different samples, pathway analysis, survival curves, etc.
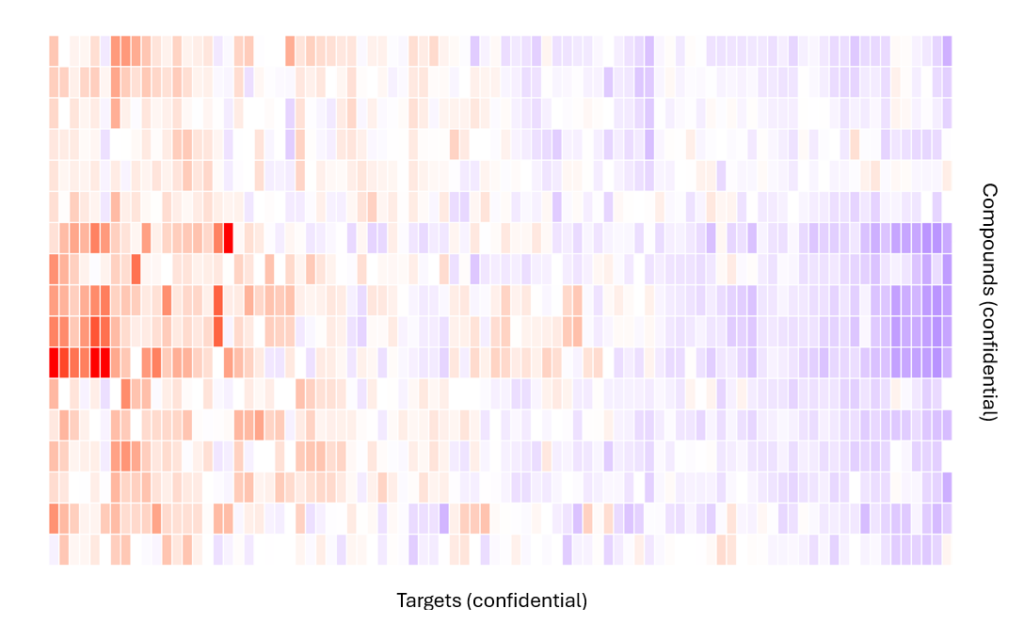
Development of an automated data processing pipeline
Creation of a script to process a large fluorescence dataset, generating all result tables (with statistical analyses) as well as PDF files with numerous graphs. The client simply needs to run the script on a dataset to generate all the required files.